We provide expert consulting and analytical guidance to help researchers and organizations implement spatial cell biology in situ approaches from concept to biological insight. Specifically, we support:
- Experimental design of in situ strategies, including selection of appropriate markers, cell types, developmental stages, and stress conditions
- Method implementation and optimization, from sample preparation and in situ protocols to imaging and quantitative acquisition
- Integration of molecular, spatial, and phenotypic data, linking gene expression, chromatin state, protein localization, and cell geometry within intact tissues
- Data analysis and interpretation, translating complex spatial datasets into biologically meaningful, cell-type–specific mechanisms
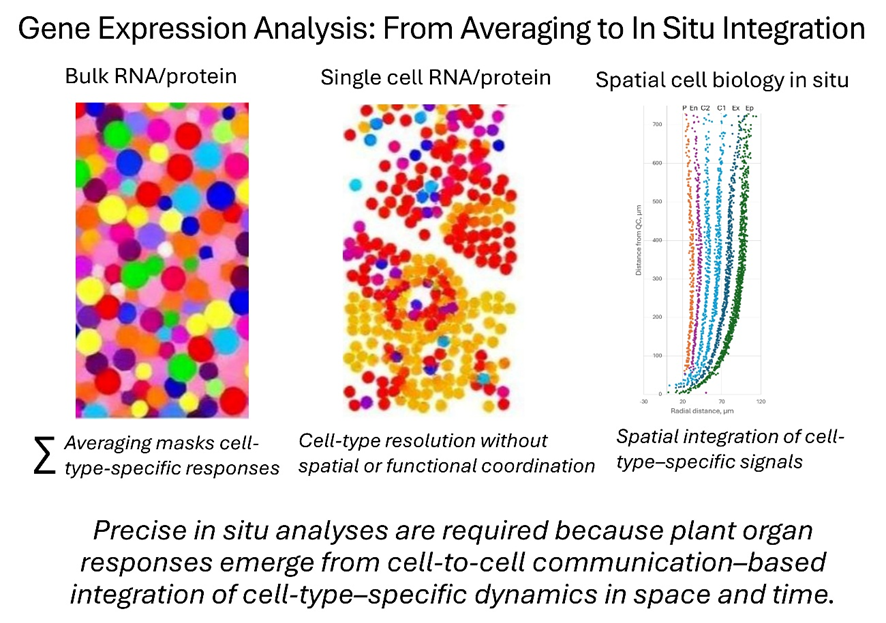
Our goal is not to replace existing molecular methods, but to embed them within a spatial, cell-type–aware framework—so that true biological mechanisms emerge from coordinated cell-to-cell interactions rather than being inferred from population averages.
| Method | Main Advantages | Key limitations | Resolution | Citations |
| Bulk RNA-seq. | Simple, cost-effective, widely used; robust for global transcriptional changes | Averages signals across all cell types; masks heterogeneity, gradients, and cell–cell interactions | Whole plant/organ/tissue-level, no spatial and single-cell resolution | Wang, 2024 Degen & Medo, 2025 |
| scRNA-seq / snRNA-seq | Resolves heterogeneity; identifies rare cell types; developmental trajectories | Protoplast/nuclear isolation stress; loss of spatial context; data sparsity, strong batch effects | Single-cell or nucleus, but dissociated from tissue | Rich-Griffin et al., 2020; Lee et al., 2025 |
| Spatial transcriptomics | Maps gene expression to tissue context; preserves partial positional gradients | Resolution is often multicellular; no chromatin or protein activity | Spatial (10–100 µm spots), transcriptome only | Giolai et al., 2 019; Li et al., 2024 Lee et al., 2025 |
| MALDI-IMS | Label-free mapping of metabolites, lipids, proteins in tissue; partially preserves distribution | No chromatin info; limited subcellular resolution; cannot capture dynamics or cell–cell interactions | Spatial (10–50 µm), metabolite/protein level | Vats et al., 2024; Shiono et al., 2024 |
| Spatial cell biology in situ | Integrates 3D organ imaging, chromatin architecture, cell/nuclear mechanosensing, cell geometry, protein localization (e.g., PLA) in organ coordinate system | Technically complex; high data integration demands | True multi-layer in situ resolution (3D chromatin, cell geometry, developmental gradients, protein interactions) | Shaw et al., 2021; Pasternak et al., 2015; Pasternak&Perez, 2021; Caballero et al., 2024 |
Key references
- Caballero, L., Pasternak, T., Riyazuddin, R., & Pérez-Pérez, J. M. (2024). Connecting high-resolution 3D chromatin maps with cell division and cell differentiation at the root apical meristem. Plant Cell Reports, 43(10), 232.
- Pasternak, T., & Pérez-Pérez, J. M. (2021). Methods of in situ quantitative root biology. Plants, 10(11), 2399.
- Teale, W. D., Pasternak, T., Dal Bosco, C., Dovzhenko, A., Kratzat, K., Bildl, W., … & Palme, K. (2021). Flavonol‐mediated stabilization of PIN efflux complexes regulates polar auxin transport. The EMBO journal, 40(1), e104416.
- Pasternak T., Falk T., Paponov I. (2020) Deep-resolution plant phenotyping platform description. dx.doi.org/10.17504/protocols.io.brsdm6a6
- Pasternak, T., Tietz, O., Rapp, K., Begheldo, M., Nitschke, R., Ruperti, B., & Palme, K. (2015). Protocol: an improved and universal procedure for whole-mount immunolocalization in plants. Plant methods, 11(1), 50.
- Pasternak T., Yaroshko O. (2026) Molecular Biology Needs a Map: Spatial In Situ Approaches in Plant Science. Plant Biology, accepted

